The GAG-DB database contains the 3D structures of GAG polysaccharides, and protein - GAG oligosaccharides. GAGs analogs and/or mimetics are also present. The following options for searching are available: 1. Presentation 2. Starting 3. Searching by fields 4. Scrolling Through the whole databaseWhat is GAG-DB ?
This database contains the three-dimensional structures of Glycosaminoglycan (GAG) binding proteins that have been crystallized with their ligands. The type of proteins, along with the bound GAG are provided. Links are available to access the information about the original article (Medline), the protein sequence, related structural information (Swissprot, PDB) ....
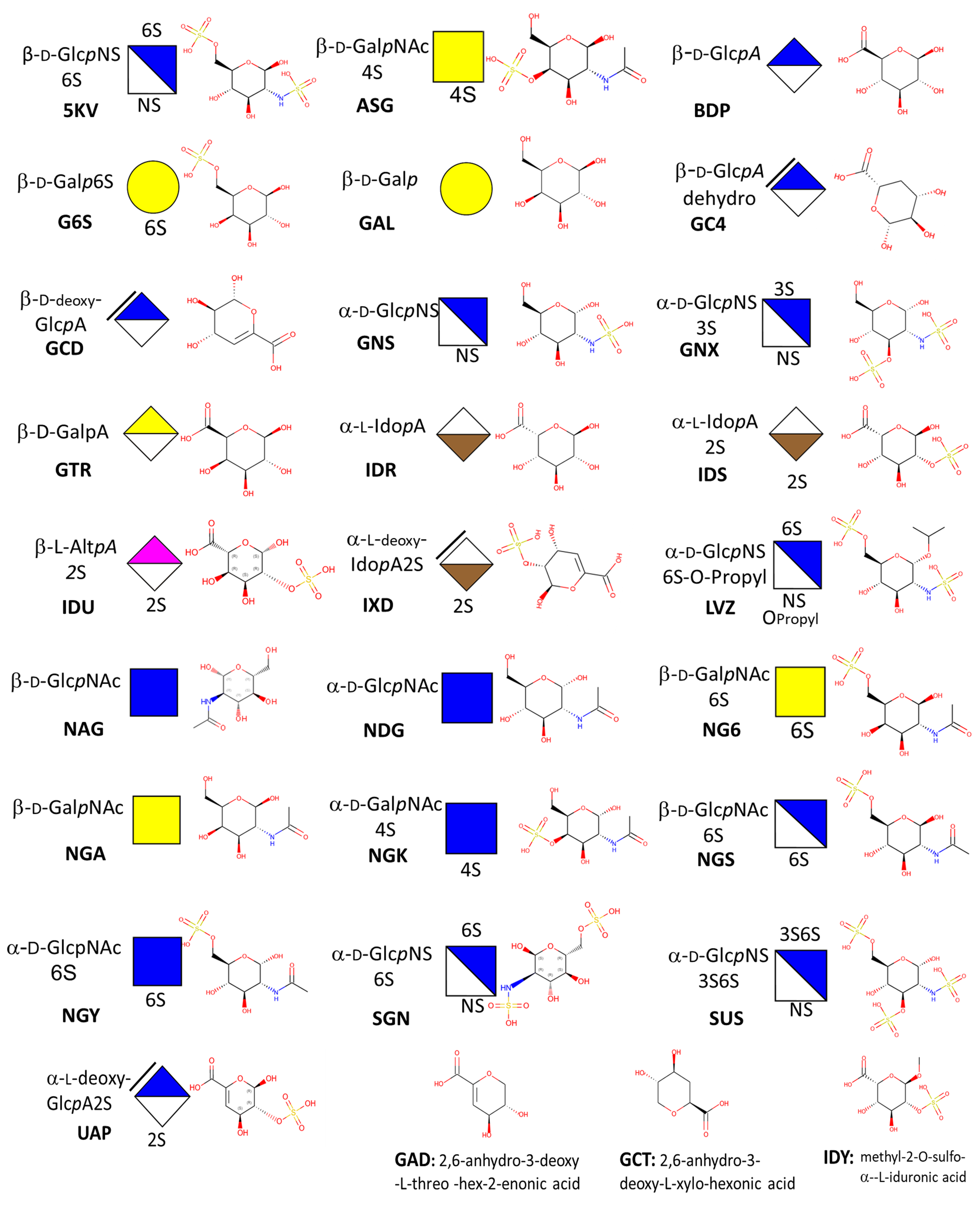
What are GAGs ?Glycosaminoglycans are linear, anionic polysaccharides, (GAGs) consisting of repeating disaccharides. Indeed, most GAG chains are formed from repeating disaccharide units of hexosamine and hexuronic acid. The exception is keratan sulfate, whose building blocks consist of hexosamine and galactose. Differences in the structure of the primary disaccharide unit regarding types of uronic acid and hexosamine, the number and position of the sulphate residues, the presence of N-acetyl and/or N-sulphate groups and the relative molecular mass. All such differences bestow these biomolecules' impressive complexity and diversity. The fine structure of the disaccharide units defines the types of GAGs. These include chondroitin/dermatan sulfate (CS/DS), heparin/heparan sulfate (Hep/HS), keratan sulfate (KS) as well as the non-sulfated hyaluronan (HA). GAGs are ubiquitously localized throughout the extracellular matrix (ECM) and to the cell membranes of cells in all tissues. They are either conjugated to protein cores in the form of proteoglycans, e.g., CS/DS, HS, and KS or as free GAGs (HA and Hep). Through their interaction with proteins, GAGs can affect cell-ECM and cell-cell interactions finely modulating ligand-receptor binding and thus chemokine and cytokine activities as well as growth factor sequestration. Indeed, it is well established that GAGs participate in the regulation of all biological processes under homeostasis; they also participate in disease progression.
PDB: PDB Code UniProt: UniProt Accession Code GlyToucan: GlyToucan Identification of the Glycan Organism: Origin of the protein or of GAG polysaccharide Function / Process / Cellular Compartiment : Biological role in compliance with Gene Ontology terms Resolution: Value in Angstrom of the crystal resolution. (set to 3 for fiber; to 0 otherwise) Gag_name: Name of the GAG Gag_max_length: Number of constituting monosaccharides Macromolecule-name: Protein or Polysaccharide or Virus name PDB_name: Title of the article as it appears in the PDB Method: Crystal Xray, Fiber Xray, NMR, Scattering, Modeling. PDB-ligand: Number and type of monosaccharide (3 letters PDB codes) Gag-mass: Molecular Mass of the GAG Pubmed: linked to publication Linucs: GAG nomenclature following (Linear Notation for Unique description of Carbohydrate Sequences For each structure a detailed page is available with 3D visualization, interactions and links to external databases see below.
On the homepage, click on "Easy Search for GAGs".
By default all available GAGs are displayed. Clicking on any of the panels will display the existing content. The user is invited to select the entry of his/her choice. This is a multi-field selection process aiming at searching the structure and directing the user directly to access the desired target. This is achieved by clicking on "Scrolling through the database".
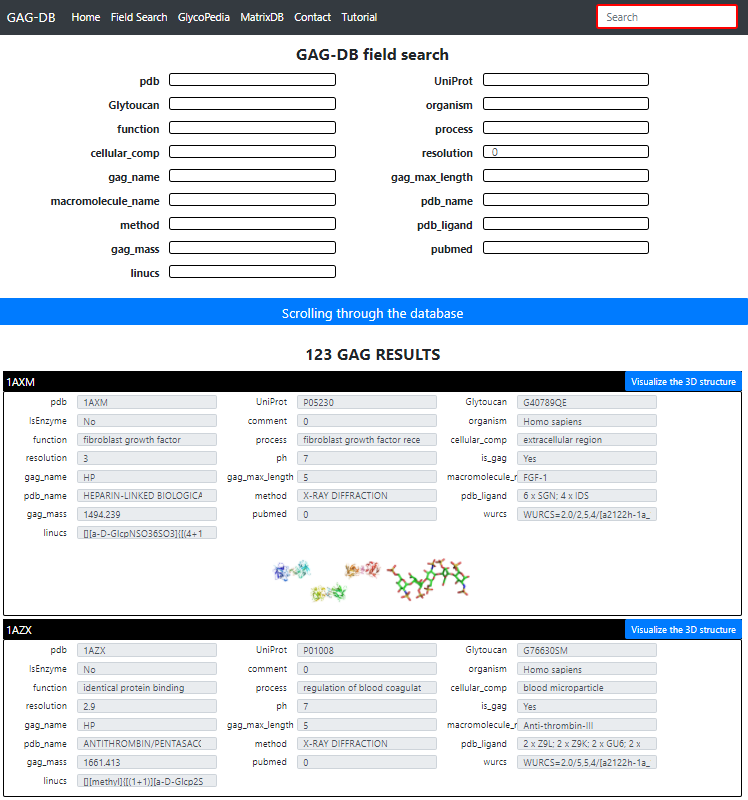
Clicking on "visualize the 3D structure" yields to the detailed page Multiple information are available for the selected GAG.

Starting again from the homepage, click on "Easy Search for GAGs". By default all available GAGs are displayed.